
dbWFA
dbWFA: A Wheat Functional Annotation Database
The search for homologous sequences and orthologs is very useful for analyzing biological sequences, especially when studying a species whose genome is still to be entirely sequenced such as wheat. Steady growth of the size of genomic databases makes homologous sequence analysis very time-consuming. Moreover, the functional annotation of genes based on their sequence homology with model species genomes requires querying unrelated databases. We developed an open-access database relating the Full-length cDNA sequences from TriFLDB TriFLDB , the transcripts of the wheat set UniGene (builds # 55,58, 59 and 60; UniGene ) and the wheat Transcription Factor DataBase (wDBTF ) that have been used to design a wheat NimbleGen 40k UniGene microarray (ArrayExpress: ref. A-MEXP-1928 ) to Arabidopsis thaliana (TAIR10 ), Oryza sativa (rice plantbiology MSU ) and Zea mays (MaizeGDB ) databases through BLAST results.
Download
The database can be downloaded freely from the dbWFA download page
Reference
Jonathan Vincent, Zhanwu Dai, Catherine Ravel, Frédéric Choulet, Said Mouzeyar, M. Fouad Bouzidi, Marie Agier and Pierre Martre dbWFA: a web-based database for functional annotation of Triticum aestivum transcripts. Database, Vol. 2013, Article ID bat014, doi:10.1093/database/bat014
Description
The 17,541 Full-Length Coding DNA Sequences from TriFLDB, which are key resources that provide a link between the genome, the transcriptome, and the proteome, as well as the 40,349, 36,692, 56,947 and 56,954 transcript sequences from the wheat UniGene set builds # 55,58, 59 and 60, respectively, which are are widely used in the genomic research community were processed using BLASTx algorithm against thaliana, Oryza sativa and Zea mays whole genomes as protein sequences (see figure below). Considering loosely that BLAST results with an e-value greater than 10-3 could not represent a suitable match for any study, they were not stored in the database. No other filter was applied to the results before their addition into the database.
The database contains Gene Ontology , MIPS A. thaliana functional classification (MAtDB ), TAIR gene families (TAIR ), PlantCyc (version 6.0) metabolic pathways and EC reactions (PMN ), and wheat transcription factors (wDBTF ). The database also contains bin information to create mapping files for the -omic data viewer and analyzing tool MapMan (BinTree version 1.1). Hand curated transcripts of wheat homologues of E3 ubiquitin ligases of the ubiquitin-proteasome system and hormone-responsive genes were also included. E3 ligases and hormone-responsive UniGenes were retrieved from the NCBI (NCBI BLAST ) and TAIR (TIGR ) databases using all Arabidopsis thaliana and Oryza sativa E3 ligases and hormone-responsive sequences as the query in a homology search using the BlastN, BlastX and TBlastX programs. The blast hits were filtered using an e-value threshold of 10-5 and an alignment length exceeding 80 bp. All sequences were checked for consistency and for the presence of specific protein signatures using InterProScan program.
Following the recommendations of the International Wheat Genome Sequencing Consortium (IWGSC ) for homology research, the percentages of coverage and identity is used to assign functional annotations to wheat transcripts. According to the IWGSC guidelines for annotating wheat genomic sequences, transcripts with a coverage and an identity higher than 50% and 45%, respectively, can be annotated with a putative function. Transcripts with a coverage and an identity higher than 90% can be annotated with a known function.
Data are stored in a MySQL database (MySQL ). The integration of the database allows one to assign the functional annotation from any of these systems to the transcripts of interest and vice versa.
Simplified database schema
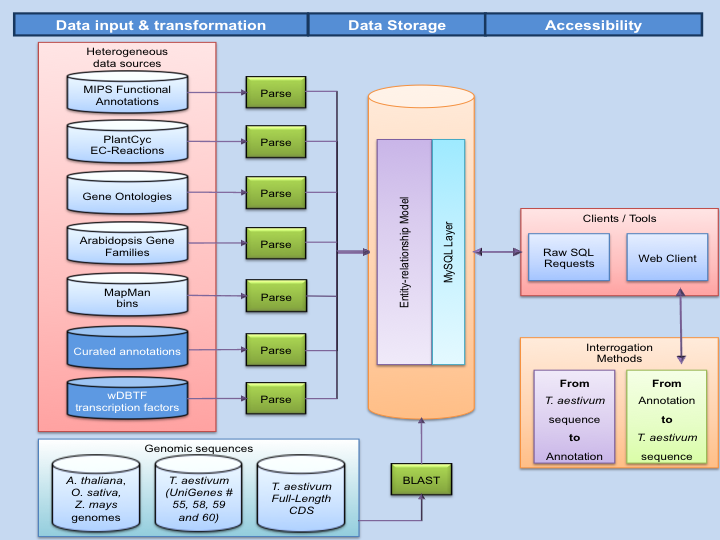
Database Structure and Website
The database contains information about Gene Ontologies , functional annotations , gene families and metabolic pathways . More than four million BLAST results were stored, allowing an efficient use of the database. Following the recommendation of the International Wheat Genome Sequencing Consortium (IWGSC ) for homology research, the percentages of coverage and identity is used to assign functional annotations to wheat transcripts.
The database was built so that one can use two different query’s approaches. From wheat transcripts IDs or NimbleGen gene expression array probes to putative family, ontology, function, and/or metabolic pathway through BLAST results, and vice versa. BLAST results allow one to characterize the homology and assign putative functional annotation between two sequences based on coverage and identity threshold values specified by the user.
A tool was developed by the "Integrative biology of grain composition team from INRA UMR1095 GDEC " in order to grant users with the most common queries that can be applied to the database. It includes searching for genes involved in specified metabolic pathways, functional annotations, gene ontologies or belonging to specified families. Or inversely, searching for putative annotations related to a wheat transcript or a list of wheat transcripts. Results are available as detailed web/html pages or text files.
Notice that this web tool is no more available, but you can download the database .
Contributors
Jonathan Vincent, ZhanWu Dai, Catherine Ravel, Fréderic Choulet, Pierre Martre,
INRA – Blaise Pascal University, UMR1095 GDEC, France
Marie Agier,
Blaise Pascal University – CNRS, UMR6158 LIMOS, France
Said Mouzeyar, M. Fouad Bouzidi,
Blaise Pascal University – INRA, UMR1095 GDEC, France
Funding
This work was supported by a Ph.D. grant from the French Ministry for Higher Education and Research to Jonathan Vincent.
Other references:
Rodrigo A. Gutiérrez, Dennis E. Shasha, and Gloria M. Coruzzi Systems Biology for the Virtual Plant. Plant Physiol., 138:550–554, 2005.
Altschul SF, Gish W, Miller W, Myers, EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol., 215:403–410, 1990.
Romeuf I, Tessier D, Dardevet M, Branlard G, Charmet G, Ravel C. wDBTF: an integrated database resource for studying wheat transcription factor families. BMC Genomics, 11:185, 2010.
Rustenholz C, Choulet F, Laugier C, Safar J, Simkova H, Dolezel J, Magni F, Scalabrin S, Cattonaro F, Vautrin S, Bellec A, Bergès H, Feuillet C and Paux E (2011) A 3000-loci transcription map of chromosome 3B unravels the structural and functional features of gene islands in hexaploid wheat. Submittedfor publication.
Mewes HW, Ruepp A, Theis F, Rattei T, Walter M, Frishman D, Suhre K, Spannagl M, Mayer KF, Stümpflen V, Antonov A. MIPS: curated databases and comprehensive secondary data resources in 2010. Nucleic Acids Res., 39:D220-4, 2011.
International Wheat Genome Sequencing Consortium. Guidelines for annotating wheat genomic sequences: release 1. Available at <http://www.wheatgenome.org/content/download/794/8948/file/wheat_gene_annotation_Release1-1.pdf>, 2006.
Romeuf I. Identification in silico des facteurs de transcription du blé tendre (Triticum aestivum) et mise en évidence des facteurs de transcription impliqués dans la synthèse des protéines de réserve. Ph.D. thesis, Université Blaise Pascal, Clermont-Ferrand, France, 2010.
Creation date: 11 Aug 2011
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use








